As light dims and food sources are limited, key changes in proteins occur in cyanobacteria
Identification of redox-sensitive enzymes can enrich biofuel production research
Using a targeted chemical biology approach, scientists at Pacific Northwest National Laboratory (PNNL) identified an important subset consisting of more than 300 proteins in a bacterium adept at converting carbon dioxide into other molecules of interest to energy researchers. These proteins are involved in generating macromolecule synthesis and carbon flux through central metabolic pathways and may also be involved in cell signaling and response mechanisms.
The team’s research also suggests that dynamic redox changes in response to specific nutrient limitations, including carbon and nitrogen limitations, contribute to the regulatory changes driven by a shift from light to dark.
They also observed that the number of labeled proteins under nitrogen or carbon limitation was ~50 percent greater than in nutrient-replete cultures, suggesting that nitrogen or carbon limitation results in increased probe labeling of proteins, indicative of a more reduced cellular environment.
“Together, our results contribute to a high-level understanding of post-translational mechanisms that regulate flux distributions and suggest potential metabolic engineering targets for redirecting carbon toward biofuel precursors,” said Dr. Charles Ansong, PNNL scientist and co-first author of the research publication that appears in Frontiers in Microbiology. “Our identification of redox-sensitive enzymes involved in these processes can potentially enrich the experimental design of research in biofuel production.”
Why It Matters: Plants or organisms that use sunlight to convert inorganic materials to organic ones for chemical compound production and respiration, among other functions, are called phototrophs. Scientists are interested in them because their conversion properties could translate into research experiments for biofuel production. But a key step toward such research is understanding protein redox chemistry-a reaction that can alter protein structure thereby regulating function. The lack of such understanding is a major void in knowledge about photoautotrophic system regulation and signaling processes.
To decrease that void, the PNNL team analyzed redox-sensitive proteins in live Synechococcus sp. PCC 7002 cells in both light and dark periods, and to understand how cellular redox balance is disrupted during nutrient perturbation.
Methods: Post-translational modifications in proteins provide a mechanism to rapidly regulate catalytic activity, and redox reactions in cysteine residues are a known mechanism. To determine the dynamic nature of the changes in redox status in Synechococcus, the scientists used cysteine thiol-specific probes, which are permeable to cells and can therefore identify redox-sensitive proteins in vivo.
They showed that this approach can detect redox changes in proteins involved in all major biological processes in Synechococcus, including proteins involved in energy metabolism, translation/protein synthesis, and photosynthesis and respiration, confirming and extending their previous observations.
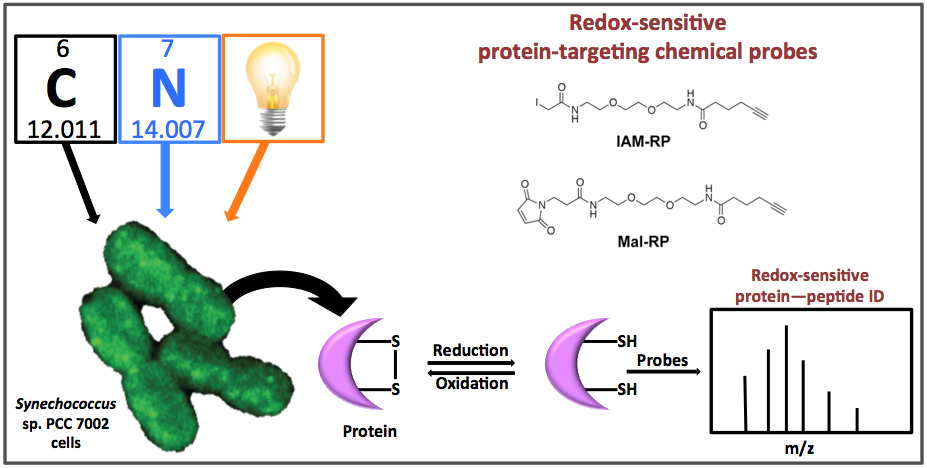
Overview of the chemical biology technique used by PNNL scientists to determine Synechococcus sp. PCC 7002 cells’ protein redox status in real time and identify redox-sensitive proteins as they occurred under induced nutrient perturbations including C and N limitation and transition from light to dark environments. Cell-permeable chemical probes derived from iodoacetamide (IAM-RP) and n-ethylmaleimide (Mal-RP) (top right) were applied to living cells. Once applied, the chemical probes irreversibly labeled proteins with reduced cysteines (bottom middle). The probe-labeled proteins were subsequently isolated for identification by high-resolution LC-MS (bottom right).
What’s Next? “Follow-up studies could include our results to match redox partners and their roles in metabolism to optimize their proposed mutants,” said Natalie Sadler, co-first author of the publication.
An example of a metabolic process well known to be redox regulated is the Calvin-Benson cycle. The scientists identified nine of the 11 Calvin-Benson enzymes providing excellent proof of concept; they now aim to pursue proteins identified by the probe that may either act as regulators or, alternatively, are regulated by redox reactions and play roles in biofuel-relevant pathways.
Acknowledgments
Sponsors: This research was supported by the Genomic Science Program (GSP) of the U.S. Department of Energy (DOE) Office of Biological and Environmental Research (BER), and is a contribution of the PNNL Biofuels and Foundational Scientific Focus Areas and the GSP-supported Pan-omics project at PNNL; mass spectrometry-based measurements and microscopy were performed in EMSL, a DOE Office of Science User Facility sponsored by BER and located at PNNL.
Research Team: Charles Ansong, Natalie C. Sadler, Eric A. Hill, Michael P. Lewis, Erika M. Zink, Richard D. Smith, Alexander S. Beliaev, Allan E. Konopka, and Aaron T. Wright, all PNNL.
User Facility: EMSL
Reference: Ansong C*, NC Sadler*, EA Hill, MP Lewis, EM Zink, RD Smith, AS Beliaev, A Konopka, and AT Wright. 2014. “Protein Redox Dynamics During Light-to-Dark Transitions in Cyanobacteria and Impacts Due to Nutrient Limitation.” Frontiers in Microbiology Online 03 July 2014. DOI: 10.3389/fmicb.2014.00325.
*Co-first authors
Source: Pacific Northwest National Laboratory, press release, 2014-09.
